# Chek if a given cycle in the graph is Hamiltonian cycle or not : O(n)
# Here, we don't run this function.
def is_hamiltonian_cycle(adj_matrix, cycle):
n = len(adj_matrix) # get number of vertices
# Check cycle length: must visit all vertices and return to start
if len(cycle) != n + 1:
return False
# Check starts and ends at the same vertex
if cycle[0] != cycle[n]:
return False
# Check all vertices (except last) are unique and cover all vertices
visited = set(cycle[:n])
if len(visited) != n:
return False
# Check edges between consecutive vertices
for i in range(n):
s, t = cycle[i], cycle[i + 1]
if adj_matrix[s][t] == 0: # No edge between u and v
return False
return True
# Convert adjacency matrix to adjacency List
def matrix_to_adj_list(adj_matrix):
adj_list = {}
n = len(adj_matrix)
for i in range(n):
adj_list[i] = set()
for j in range(n):
if adj_matrix[i][j] == 1:
adj_list[i].add(j)
return adj_list
# for adjacency List (We use this one!)
def is_hamiltonian_cycle_list(adj_list, cycle):
n = len(adj_list) # number of vertices
if len(cycle) != n + 1:
return False
if cycle[0] != cycle[n]:
return False
visited = set(cycle[:n])
if len(visited) != n:
return False
for i in range(n):
s, t = cycle[i], cycle[i + 1]
if t not in adj_list[s]:
return False
return True
# Convert the vertex numbers (0, 1, ...,) to letters (A, B, ...,)
def print_adj_list(adj_list):
print("Adjacency List: ")
for v in adj_list:
v_char = chr(ord('A') + v)
neighbors = [chr(ord('A') + u) for u in adj_list[v]]
neighbors_str = ', '.join(sorted(neighbors))
print(f"{v_char}: {neighbors_str}")
print()
# Print cycle and its result
def print_cycle_with_result(cycle, adj_list):
cycle_letters = [chr(ord('A') + v) for v in cycle]
print("Cycle: ", " → ".join(cycle_letters))
is_hamiltonian = is_hamiltonian_cycle_list(adj_list, cycle)
print("Is this Hamiltonian?:", is_hamiltonian)
print()
if __name__ == "__main__":
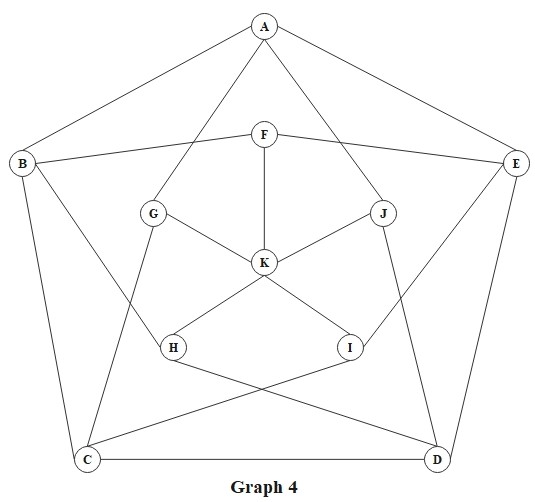
# Example: Graph 4
G_4 = [
[0, 1, 0, 0, 1, 0, 1, 0, 0, 1, 0], # Vertex A
[1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0], # Vertex B
[0, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0], # Vertex C
[0, 0, 1, 0, 1, 0, 0, 1, 0, 1, 0], # Vertex D
[1, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0], # Vertex E
[0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1], # Vertex F
[1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1], # Vertex G
[0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1], # Vertex H
[0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1], # Vertex I
[1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1], # Vertex J
[0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 0], # Vertex K
]
# Use the adjacency List
adj_list_4 = matrix_to_adj_list(G_4)
print_adj_list(adj_list_4)
# Valid Hamiltonian cycle
cycle1 = [0, 6, 2, 8, 10, 7, 1, 5, 4, 3, 9, 0]
print_cycle_with_result(cycle1, adj_list_4)
# Invalid cycle
cycle2 = [0, 1, 2, 3, 4, 5, 7, 6, 10, 9, 8, 0]
print_cycle_with_result(cycle2, adj_list_4)